
 Based on these models, a great number of ab initio gene prediction programs have been developed. Many algorithms are applied for modeling gene structure, such as Dynamic Programming, linear discriminant analysis, Linguist methods, Hidden Markov Model and Neural Network. The search by this method thus relies on the major feature present in the genes. Exon detection must rely on the content sensors. On the other hand content sensors refer to the patterns of codon usage that are unique to a species, and allow coding sequences to be distinguished from the surrounding non-coding sequences by statistical detection algorithms. Signal sensors refer to short sequence motifs, such as splice sites, branch points, polypyrimidine tracts, start codons and stop codons.
Based on these models, a great number of ab initio gene prediction programs have been developed. Many algorithms are applied for modeling gene structure, such as Dynamic Programming, linear discriminant analysis, Linguist methods, Hidden Markov Model and Neural Network. The search by this method thus relies on the major feature present in the genes. Exon detection must rely on the content sensors. On the other hand content sensors refer to the patterns of codon usage that are unique to a species, and allow coding sequences to be distinguished from the surrounding non-coding sequences by statistical detection algorithms. Signal sensors refer to short sequence motifs, such as splice sites, branch points, polypyrimidine tracts, start codons and stop codons. 
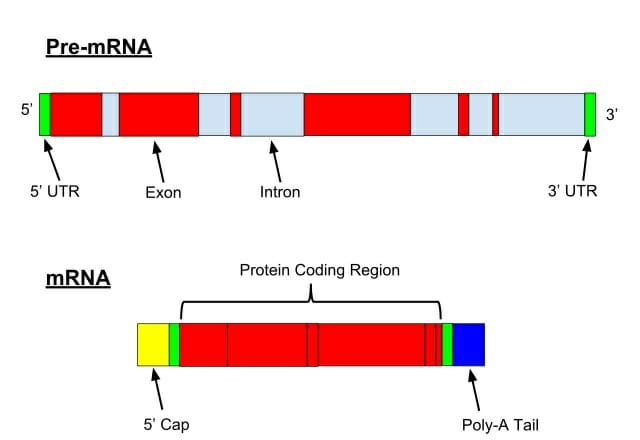
Ab initio gene predictions rely on two types of sequence information: signal sensors and content sensors.It uses gene structure as a template to detect genes.It is a method based on gene structure and signal-based searches. A new heuristic method based on pairwise genome comparison has been implemented in the software called CSTfinder.Two more types of software, PROCRUSTES and GeneWise , use global alignment of a homologous protein to translated ORFs in a genomic sequence for gene prediction.The most common local alignment tool is the BLAST family of programs, which detects sequence similarity to known genes, proteins, or ESTs.
 Local alignment and global alignment are two methods based on similarity searches. Once there is similarity between a certain genomic region and an EST, DNA, or protein, the similarity information can be used to infer gene structure or function of that region. This approach is based on the assumption that functional regions (exons) are more conserved evolutionarily than nonfunctional regions (intergenic or intronic regions). It is a conceptually simple approach that is based on finding similarity in gene sequences between ESTs (expressed sequence tags), proteins, or other genomes to the input genome. It is a method based on sequence similarity searches.
Local alignment and global alignment are two methods based on similarity searches. Once there is similarity between a certain genomic region and an EST, DNA, or protein, the similarity information can be used to infer gene structure or function of that region. This approach is based on the assumption that functional regions (exons) are more conserved evolutionarily than nonfunctional regions (intergenic or intronic regions). It is a conceptually simple approach that is based on finding similarity in gene sequences between ESTs (expressed sequence tags), proteins, or other genomes to the input genome. It is a method based on sequence similarity searches.







 0 kommentar(er)
0 kommentar(er)
